Inhalt
Influenza and Arterivirus:
The role of membrane proteins for cell entry and exit of viruses
Influenza virus
The function of HA and M2 during virus assembly and budding
The glycoprotein hemagglutinin (HA) of influenza virus catalyses membrane fusion, but is also the driving force for budding of virus particles from the plasma membrane. Concentration of HA in membrane-rafts, microdomains of the plasma membrane enriched in cholesterol and sphingolipids, serves as a platform for assembly of the other viral proteins. Funded in the framework of a DFG priority program (link), we have characterized the interactions of HA with lipid domains essentially using biophysical methods, both in living cells (FRET, FRAP) and in model membranes. We determined the signals for raft-association, hydrophobic amino acids in the transmembrane region and S-acylation of cytoplasmic cysteines. In another DFG-funded, Russian-German project (Ve 141/10), the molecular requirements for the attachment of different fatty acids (palmitate and stearate) to HA and the glycoprotein HEF of Influenza C virus and its functional consequences for virus budding and membrane fusion are analysed.
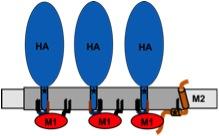
Model for assembly of influenza virus proteins in membrane-rafts.
The viral budozone (grey) is enriched in raft-lipids. Three fatty acids attached to the cytoplasmic tail of HA, two palmitate (black lines) and one stearate (red line), target the protein to the budozone. M1 (red) interacts with the cytoplasmic tail of HA, oligomerizes and forms a shell underneath the membrane. M2 (brown) localizes to the edge of the budozone by means of a palmitate and a cholesterol-binding site (star). The amphiphilic helix is believed to insert into the membrane, thereby inducing membrane curvature to mediate scission of virus particles.
M2, the proton channel of influenza virus, contains an amphiphilic helix in its cytoplasmic domain, which is palmitoylated and binds the raft-lipid cholesterol. Despite having these raft-targeting features, M2 is not associated with membrane rafts. A recent model suggests that M2 is located at the edge of rafts where it inserts its amphiphilic helix into the membrane to mediate pinching-off of virus particles. In the framework of the SFB 740 (link) we are testing this model with biophysical techniques in transfected cells as well as with reverse genetics in the context of virus replication.
Arteriviruses
Arteriviridae are a family of enveloped, positive-stranded RNA-viruses, which are poorly characterized in molecular terms. Prominent members are the equine arteritis viruses (EAV) and the porcine reproductive and respiratory syndrome viruses (PRRSV), the most important pathogen in pig breeding. Our research is focused on the structure and function of the membrane proteins, the two spike protein complexes GP2/3/4 and GP5/M, and the small E protein, a putative proton channel.
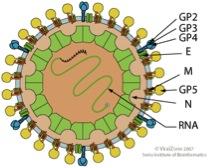
Scheme of an Arterivirus particle (from viral zone)
The function of the GP2/3/4 spike during cell entry
In the context of the EU-funded Initial Training Network Virus Entry (link), we are analysing the function of the GP2/GP3/GP4 complex of equine arteritis virus during cell entry. We have determined basic molecular features of the GP2/GP3/GP4 complex, its membrane topology and processing, such as signal peptide cleavage and glycosylation. A long term aim of the project is to express the viral spike on the plasma membrane of transfected cells to perform receptor-binding and membrane fusion assays. Another part of the project deals with the expression of a soluble version of GP2/GP3/GP4 in insect cells to purify the complex for structural analysis.
Persistent infection with PRRSV
Structural characterisation of glycoprotein 5 to analyze antibody epitopes
Due to the late appearance of neutralizing antibodies PRRSV persists in lymphoid tissues for months. Most antibodies are directed against glycoprotein 5 (GP5), which forms a disulfide-linked complex with the M-protein. Two antibody epitopes have been localized on GP5, a non-neutralizing decoy epitope A located in the predicted signal peptide and the neutralizing epitope B in the ectodomain. The epitopes are separated by a hypervariable region containing a variable number of glycosylation sites. This model is, however, not proven in its basic molecular preconditions.
Funded by the DFG (Ve 141/11) and the PRRSV-research prize 2012 from Boehringer VetMedica, we are testing GP5 from various PRRSV-strains with additions and deletions of glycosylation sites for signal peptide cleavage and the role of carbohydrates and other protein modifications for binding of antibodies.
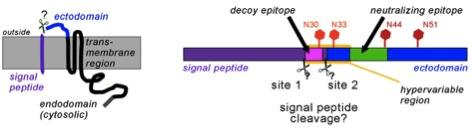
Primary structure of GP5 with the (predicted) signal peptide (purple), ectodomain (blue) and transmembrane and cytosolic region (black).The right part shows the decoy-epitope A (pink) and the neutralizing epitope B (green) within a hypervariable region. Conserved (dark red) and variable (light red) glycosylation sites are indicated by hexagons.
Links :
Wann wird es für den Menschen gefährlich?
DSFG-Datenbank Depris über die PRRSV Projekte:
http://gepris.dfg.de/gepris/projekt/256219832
http://gepris.dfg.de/gepris/projekt/193516483