Targeted sequence enrichment and sequencing of zoonotic orthohantaviruses
We study genetic diversity, evolution and pathogenicity of zoonotic orthohantaviruses which circulate in wild rodent populations in Europe. We use a combination of the targeted sequence enrichment and high-throughput sequencing to obtain full sequences of hantaviruses genomes from clinical samples. Our approach enables selective sequencing of a broad range of orthohantaviruses, including Dobrava-Belgrade, Hantaan, Puumala, Soul and Tula orthohantaviruses (see below for more detail).
Emerging diseases are a major threat to public health. Over the past two decades many different zoonotic viruses, such as Severe acute respiratory syndrome coronavirus, Middle East respiratory syndrome coronavirus, avian influenzaviruses, orthohantaviruses, Zaire ebolavirus, Zika virus, (re)emerged as human pathogens. The majority of viruses that are known to infect humans are perpetuated naturally in animals, and wild animals are the major source of these viruses.
Genetic changes fuel viral evolution and help viruses to adapt to new environments. Yet, genetic changes that underlie human adaptation, human-to-human transmissibility and pathogenesis in zoonotic viruses are only poorly understood.
Hantaviruses
Orthohantaviruses of the family Hantaviridae are important emerging viruses that can be transmitted from rodents to humans. They cause hemorrhagic fever with renal syndrome in Eurasia and hantavirus cardiopulmonary syndrome in the Americas. No specific treatment or approved vaccine is available against diseases caused by orthohantaviruses. In fact, orthohantaviruses are the most prevalent zoonotic viruses in Germany and three different orthohantaviruses – Puumala, Dobrava-Belgrade, and Tula orthohantaviruses – circulate in wild rodent populations in Germany.
In Germany, 3,663 orthohantavirus infections were diagnosed in years 2010-2015, and PUUV was responsible for 97% of the diagnosed cases. The high fluctuation of human infections correlates with the fluctuation of its natural animal reservoir, the bank vole Myodes glareolus, a dominant rodent species of European forests. While the bank vole is widely distributed in entire Germany, most of the PUUV-associated cases were recorded in several endemic regions in the south and west of the country.
In general, it is difficult to obtain clinical samples (blood samples) from human orthohantavirus patients during the acute phase of the disease. Such samples are very rare, however, they are very valuable, because they may provide a better understanding of the evolution of viruses and the discovery of genetic changes that are important for interspecies transmission. Physicians, suspecting a orthohantavirus infection are instructed to contact the National Consultant Laboratory for Hantaviruses at the Institute for Virology of Charité-Universitätsmedizin Berlin (https://virologie-ccm.charite.de/diagnostik/konsiliarlaboratorium_fuer_hantaviren/). The consultant laboratory offers accurate virus diagnostics tests and provides expert advice on infection epidemiological questions.
Primarily, we investigate the genetic make-up of Puumala, Dobrava-Belgrade and Tula Orthohantaviruses. We seek to understand how these viruses evolve and what genetic markers are important for their pathogenicity and transmission to new host species.
Targeted sequence enrichment & sequencing
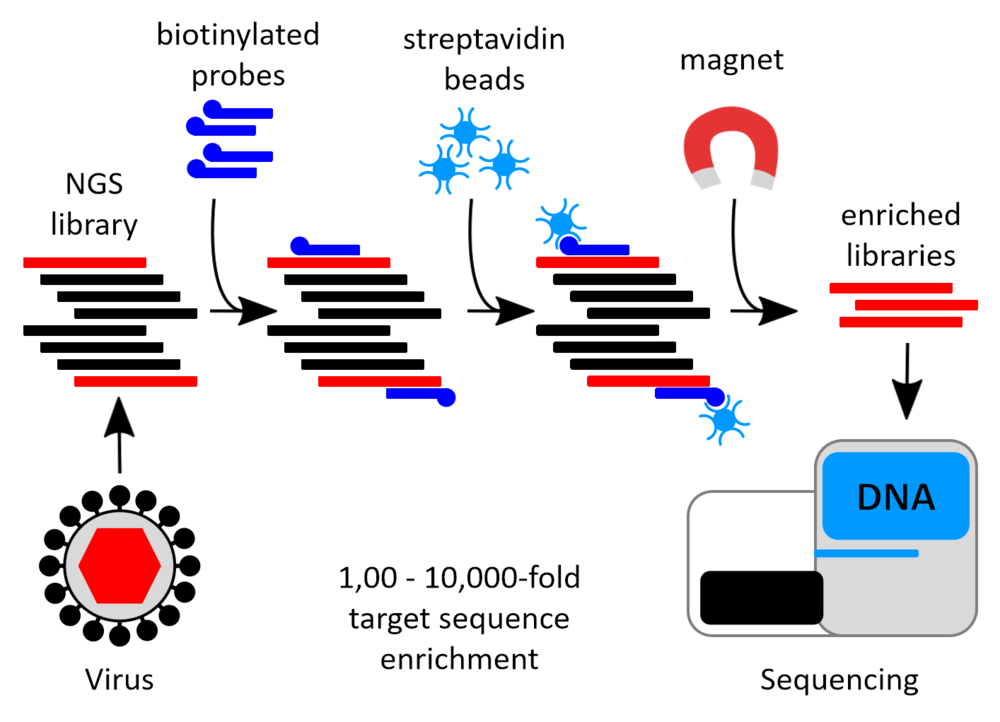
We use a combination of the targeted sequence enrichment and high-throughput sequencing to obtain complete genomic sequences of various strains of orthohantaviruses directly from samples from the reservoir hosts. Targeted enrichment allows specific purification of minute amounts of nucleic acids from heterogeneous mixes, and thus to focus sequencing efforts on important targets. This approach is well applicable in real infection scenarios. We used the method to obtain complete genomic sequences from various strains of different orthohantaviruses directly from samples from the reservoir hosts.
Targeted sequence enrichment uses a pool of biotinylated oligonucleotide probes that are designed to hybridize to fragmented DNA sequences of interest. After hybridization, the captured targets are purified from the initial material using reactive, magnetic beads and sequenced on a high-throughput platform (Figure).
Currently, our approach enables selective purification and sequencing of the following orthohantaviruses:
- Dabieshan orthohantavirus
- Dobrava-Belgrade orthohantavirus
- Kurkino virus
- Saaremaa virus
- Sangassou virus
- Sochi virus
- Hantaan orthohantavirus
- Amur virus
- Soochong avirus
- Puumala orthohantavirus
- Seoul orthohantavirus
- Thailand orthohantavirus
- Anjozorobe hantavirus
- Jurong hantavirus
- Serang hantavirus
- Tula orthohantavirus
If you have samples containing any of the above mentioned viruses and are interested in an academic collaboration, please get in touch and we can discuss possible opportunities.